Class ExponentialMovingFeature
Import
from NitroFE import ExponentialMovingFeature
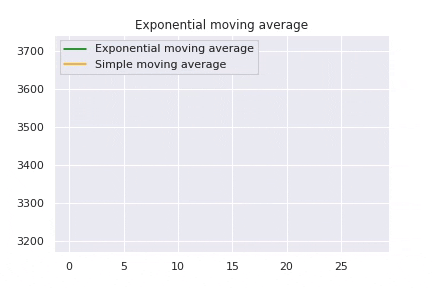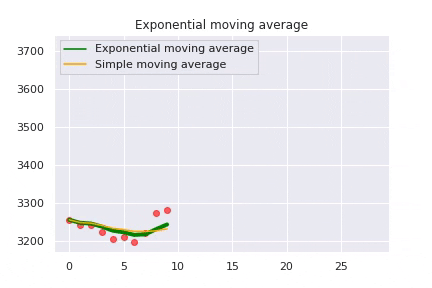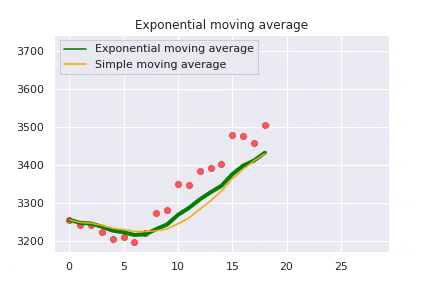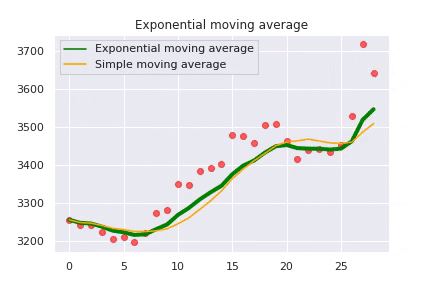
The exponential moving average is caluclated as
if you want you calculate via the traditional way, in which the ema[0] isnt the first value in the dataframe (usually a simple moving average over the first few values ),
you can use the paramters 'initialize_using_operation' and 'initialize_span' , in which case the exponential moving avergae will be calculated as
Methods
Provided dataframe must be in ascending order.
__init__(self, alpha=None, operation='mean', initialize_using_operation=False, initialize_span=None, com=None, span=None, halflife=None, min_periods=0, ignore_na=False, axis=0, times=None)
special
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
alpha |
float |
Specify smoothing factor directly, by default None |
None |
operation |
str |
operation to be performed for the moving feature,available operations are 'mean','var','std', by default 'mean' |
'mean' |
initialize_using_operation |
bool |
If True, then specified 'operation' is performed on the first 'initialize_span' values, and then the exponential moving average is calculated, by default False |
False |
initialize_span |
int |
the span over which 'operation' would be performed for initialization, by default None |
None |
com |
float |
Specify decay in terms of center of mass, by default None |
None |
span |
int |
specify decay in terms of span , by default None |
None |
halflife |
float |
Specify decay in terms of half-life, by default None |
None |
min_periods |
int |
Minimum number of observations in window required to have a value (otherwise result is NA), by default 0 |
0 |
ignore_na |
bool |
Ignore missing values when calculating weights; specify True to reproduce pre-0.15.0 behavior, by default False |
False |
axis |
int |
The axis to use. The value 0 identifies the rows, and 1 identifies the columns, by default 0 |
0 |
times |
str |
Times corresponding to the observations. Must be monotonically increasing and datetime64[ns] dtype, by default None |
None |
Source code in nitrofe\time_based_features\moving_average_features\moving_average_features.py
def __init__(
self,
alpha: float = None,
operation: str = "mean",
initialize_using_operation: bool = False,
initialize_span: int = None,
com: float = None,
span: int = None,
halflife: float = None,
min_periods: int = 0,
ignore_na: bool = False,
axis: int = 0,
times: str = None,
):
"""
Parameters
----------
alpha : float, optional
Specify smoothing factor directly, by default None
operation : str, {'mean','var','std'}
operation to be performed for the moving feature,available operations are 'mean','var','std', by default 'mean'
initialize_using_operation : bool, optional
If True, then specified 'operation' is performed on the first 'initialize_span' values, and then the exponential moving average is calculated, by default False
initialize_span : int, optional
the span over which 'operation' would be performed for initialization, by default None
com : float, optional
Specify decay in terms of center of mass, by default None
span : float, optional
specify decay in terms of span , by default None
halflife : float, optional
Specify decay in terms of half-life, by default None
min_periods : int, optional
Minimum number of observations in window required to have a value (otherwise result is NA), by default 0
ignore_na : bool, optional
Ignore missing values when calculating weights; specify True to reproduce pre-0.15.0 behavior, by default False
axis : int, optional
The axis to use. The value 0 identifies the rows, and 1 identifies the columns, by default 0
times : str, optional
Times corresponding to the observations. Must be monotonically increasing and datetime64[ns] dtype, by default None
"""
self.com = com
self.span = span
self.halflife = halflife
self.alpha = alpha
self.min_periods = min_periods if min_periods != None else 0
self.adjust = False
self.ignore_na = ignore_na
self.axis = axis
self.times = times
self.operation = operation
self.last_values_from_previous_run = None
self.initialize_using_operation = initialize_using_operation
self.initialize_span = initialize_span
fit(self, dataframe, first_fit=True)
For your training/initial fit phase (very first fit) use fit_first=True, and for any production/test implementation pass fit_first=False
Parameters:
| Name | Type | Description | Default |
|---|---|---|---|
dataframe |
Union[pandas.core.frame.DataFrame, pandas.core.series.Series] |
dataframe containing column values to create exponential moving feature over |
required |
first_fit |
bool |
Moving features require past values for calculation. Use True, when calculating for training data (very first fit) Use False, when calculating for subsequent testing/production data { in which case the values, which were saved during the last phase, will be utilized for calculation }, by default True |
True |
Source code in nitrofe\time_based_features\moving_average_features\moving_average_features.py
def fit(self, dataframe: Union[pd.DataFrame, pd.Series], first_fit: bool = True):
"""
For your training/initial fit phase (very first fit) use fit_first=True, and for any production/test implementation pass fit_first=False
Parameters
----------
dataframe : Union[pd.DataFrame, pd.Series]
dataframe containing column values to create exponential moving feature over
first_fit : bool, optional
Moving features require past values for calculation.
Use True, when calculating for training data (very first fit)
Use False, when calculating for subsequent testing/production data { in which case the values, which
were saved during the last phase, will be utilized for calculation }, by default True
"""
if not first_fit:
if self.last_values_from_previous_run is None:
raise ValueError(
"First fit has not occured before. Kindly run first_fit=True for first fit instance,"
"and then proceed with first_fit=False for subsequent fits "
)
self.adjust = False
dataframe = pd.concat(
[self.last_values_from_previous_run, dataframe], axis=0
)
else:
if self.initialize_using_operation:
self.min_periods = 0
if (self.initialize_span is None) and (self.span is None):
raise ValueError(
"For initialize_using_operation=True,"
"either initialize_span or span value is required"
)
elif (self.initialize_span is None) and (self.span is not None):
self.initialize_span = self.span
first_frame = self._perform_temp_operation(
dataframe[: self.initialize_span].rolling(
window=self.initialize_span
)
)
dataframe = pd.concat([first_frame, dataframe[self.initialize_span :]])
else:
if self.initialize_span is not None:
raise ValueError(
"In order to use initialize_span, initialize_using_operation must be True"
)
_dataframe = dataframe.ewm(
com=self.com,
span=self.span,
halflife=self.halflife,
alpha=self.alpha,
min_periods=self.min_periods,
adjust=self.adjust,
ignore_na=self.ignore_na,
axis=self.axis,
times=self.times,
)
_return = self._perform_temp_operation(_dataframe)
if not first_fit:
_return = _return.iloc[1:]
self.last_values_from_previous_run = _return.iloc[-1:]
return _return